RNAi Libraries
The VDRC genome-wide Drosophila RNAi resource enables large scale genetic screens, making it possible to carry out loss-of-function experiments in essentially any tissue or cell at any stage in the life of the organism.
When crossed to a Gal4 'driver' line, the UAS-RNAi stocks induce expression of a specific hairpin structure, which silences expression of the target gene via RNA interference (RNAi).
The VDRC RNAi resource includes transgenic UAS-RNAi stocks with long hairpins (GD and KK libraries) and short hairpin microRNAs (shRNA library).
- Collectively, the RNAi lines cover a total of 12,671 (91%) of the D.melanogaster protein-coding genes.
- GD collection covers 11,292 genes (81%).
- KK collection covers 9,646 genes (69%).
- shRNA collection covers 372 genes (2.7%) - more under construction.
- For over 8500 genes, we have 2 or more independent RNAi lines available.
In addition, the VDRC provides:
- 13,848 DNA constructs used to generate the GD collection.
- Toolkit stocks used for construction of all 3 RNAi libraries.
RNAi Libraries Overview
Collectively, the RNAi lines cover a total of 12,671 (91%) of the D.melanogaster protein-coding genes. For over 8500 genes, we have 2 or more independent RNAi lines available.
| P-element library (GD) | ΦC31 library (KK) | Short hairpin microRNA library (shRNA) | |
|---|---|---|---|
| Available since | 2007 | 2009 | 2016 |
| Insertion of the UAS-IR | Random. Insertions mapped to chromosome X, 2 or 3. Precise locations unknown. | Targeted into VIE-260b landing site, chromosome 2, at position 22019296 (5' to CR33987) (see KK library - landing site information) | Targeted into attP40 landing site (Markstein et al., 2008), chromosome 2 at 25C6 |
| Target selection/ PCR template design | Dietzl et al (2007) Nature v448 p151 | Horn and Boutros (DKFZ, Heidelberg) | DSIR website tool and off-target analysis script (Burkard, IMP/IMBA, Vienna) |
| Hairpin length range/average (bp) | 109-400 (321) | 81-799 (357) | 21 |
| Transformation vector | pMF3 (10x UAS) | pKC26 (10x UAS) | pWALIUM20 (Perkins et al., 2015) |
| Verification of transformant lines | Restriction digest | Sanger sequencing | Sanger sequencing |
| Coverage | 16,763 lines 11,292 genes (81%) Average of 1.19 lines/gene |
9,822 lines 9,646 genes (69%) Average of 0.7 lines/gene |
372 lines (as of Jan 2017). More to be added. |
| Activity | Soma and germline. Enhanced by co-expression of Dicer2 | Soma and germline. Enhanced by co-expression of Dicer2 | Soma and germline. |
GD Transgenic RNAi Library
The GD library comprises 16,763 transgenic Drosophila strains, each containing an inducible UAS-RNAi construct against a single protein-coding gene. All insertions have been molecularly validated, and a sample also functionally validated. We estimate that >80% of the lines provide potent and gene-specific silencing.
GD library cloning strategy
The library was constructed by cloning gene fragments of 300-400bp as inverted repeats (IRs) in the antisense-sense orientation into a modified pUAST vector pMF3 with 10 copies of UAS sites (see map). To amplify 300-400bp gene fragments, primers were designed to amplify from every predicted protein-coding gene in Release 4.3 of the Drosophila genome sequence. 15,072 PCR products were successfully amplified and cloned as IRs into pMF3, representing 13,344 genes or 97% of the Drosophila genome. The GD library insertions are P-element based transgenes with random insertion sites. Insertions have been mapped to a chromosome (1, 2, or 3) but we do not have data on specific location on the chromosome.
Publication showing creation and validation of GD library:
- A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila.
Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ.
Nature. 2007 Jul 12;448(7150):151-6.
PMID: 17625558
[Abstract]
GD Protocols
- GD library cloning strategy
- Genomic DNA extraction
- mRNA extraction
- 1st strand cDNA
- 96 well cloning of RNAi constructs
- 96 well DNA purification using 'SigmaSpin Post-Reaction Clean-Up Plates'
- 96 well DNA gel extraction using 'NucleoSpin Multi-96 Extract'
- Fly transformation
- Bacteria Transformation
- Transgene Verification
- pMF3 transformation vector [MAP] [PDF] [TEXT]
KK Transgenic RNAi Library
The KK library, created after the GD library, is a genome-wide transgenic RNAi library.
The main motivation for generating a new library was to:
- improve consistency and viability by integrating all constructs at the same landing site
- increase efficiency and specificity of gene disruption by using improved Inverted Repeat (IR) design
- generate a line for genes not covered by the GD library
- provide an independent construct to verify phenotypic effects.
A key difference between the GD and KK libraries is that the GD library insertions are P-element based transgenes with random insertion sites, whereas the KK library contains phiC31-based transgenes with a single, defined insertion site.
The KK library currently comprises 9,822 transgenic Drosophila strains, each containing an inducible UAS-RNAi construct against a single protein-coding gene. The sequence of all insertions have been validated by sequencing, and a sample also functionally validated. Based on an initial benchmark study of 164 genes with known lethality/morphological phenotypes, >90% of the KK lines provided potent and gene-specific silencing when crossed to the ubiquitous driver Act5C-GAL4 (Keleman et al., unpublished).
KK library - landing site information
KK RNAi lines landing site - background information
During construction of the KK RNAi library, the hairpin constructs were targeted to a single genomic location. However, Green et al. (2014) have previously shown that there are actually 2 potential genomic landing sites for the KK RNAi constructs:
- At 40D3 (the originally intended VIE-260B landing site, in the 5'UTR of the gene tiptop, position chr2L: 22019296)
- At 30B3 (the actual site, position chr2L: 9437482).
In 75% of the KK lines, the insertion of the RNAi hairpin construct is in the 30B landing site only and there are no problems. However, in ~25% of KK lines there is an additional insertion of the RNAi construct at the 40D landing site and this causes non-specific phenotypes when crossed to some Gal4 drivers. See Green et al., Nat. Methods (2014) for details.
A publication investigating this issue further, from the lab of Kieran Harvey (Vissers et al., Nat. Comm. (2016)), confirms that approximately 25% of VDRC KK RNAi lines (38 out of 150) can cause false-positive phenotypes. Whilst working on the hippo pathway and performing screens in the eye, Vissers et al. noticed phenotypes which were due to the insertion site and not due to expression of the RNAi hairpin. They were able to attribute the false-positive phenotypes to enhancement of the Hippo pathway, owing to ectopic UAS-driven expression of the Tiptop (tio) transcription factor - a consequence of the 40D landing site being located in the tio 5' UTR.
KK library control lines
As a control line for experiments involving KK lines, the VDRC provides the host strain for the KK library (VDRC_ID 60100). This line contains landing sites at both 40D and 30B.
A number of additional control lines for the KK library, donated by Kieran Harvey (Peter MacCallum Cancer Centre, Melbourne, Australia) are available. (For details see Vissers et al., (2016): A Drosophila RNAi library modulates Hippo pathway-dependent tissue growth).
The collection consists of 5 lines which can be displayed by using the search term "....harvey".
An RNAi-toolkit stock, 40D-UAS (VDRC_ID 60101) where the landing sites at 40D and 30B in the KK library host strain have been separated by meiotic recombination. This line contains Gal4-responsive UAS repeats but no functional RNAi coding sequence at 40D and no transgene insertion at 30B and enables researchers to easily test, in a single cross, whether their genetic screen of interest will be affected by ectopic tiptop expression.
4 further KK lines for specific genes were created by meiotic recombination to separate the insertions at 30B and 40D. These lines contain an insertion of the UAS-RNAi construct in the landing site at 30B only and no insertion at the 40D landing site.
- VDRC_ID 111000 is the '30D only' version of VDRC_ID 107151 (CG8208, MBD-like)
- VDRC_ID 111001 is the '30D only' version of VDRC_ID 104523 (CG4005, Yki)
- VDRC_ID 111002 is the '30D only' version of VDRC_ID 106174 (CG12072, Wts)
- VDRC_ID 111003 is the '30D only' version of VDRC_ID 105838 (CG4236, Caf1)
When using these additional control lines, please reference Vissers et al (2016) and acknowledge the VDRC for providing the lines.
We would like to emphasise that ~75% of KK lines (insertion at 30D only) are absolutely fine and that the remaining 25% (insertion at 40D) will only be a problem in cases where an assay is affected by misexpression of the tio gene. If you are planning to use KK lines and are concerned that you will see non-specific phenotypes, we suggest that you cross your Gal4 driver of interest to the 40D-UAS line and check whether the F1 progeny demonstrate a dominant phenotype. If your assay is not affected by tio misexpression, you can use all 9,822 KK lines with confidence, but if you see an effect, then 25% of the KK lines will give misleading results in that particular screen.
Validation of KK lines
The VDRC have shipped more than 400,000 KK lines to the Drosophila research community. We would like to reassure researchers that use of KK lines has resulted in several hundred publications in which the KK line phenotypes have been validated with independent RNAi lines targeting a different region of the same gene and with other insertion sites. These publications include some large scale screens, e.g.:
- Czech B et al., (2013). A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol Cell. 2013 Jun 6;50(5):749-61.
[PubMed]. - Berns N et al., (2014). A genome-scale in vivo RNAi analysis of epithelial development in Drosophila identifies new proliferation domains outside of the stem cell niche. J Cell Sci. 2014 Jun 15;127(Pt 12):2736-48.
[PubMed]. - Homem CC et al., (2014). Ecdysone and mediator change energy metabolism to terminate proliferation in Drosophila neural stem cells. Cell. 2014 Aug 14;158(4):874-88.
[PubMed].
Verify your results!
As is recommended for results obtained using any RNAi lines, we advise verifying your observed KK phenotype with an alternative RNAi line containing an independent RNAi hairpin construct targeting the same gene and a different genomic insertion site where possible.
Please note that the GD library was generated using P-element insertion and the shRNA library transgenes are inserted in the attP40 landing site, therefore the landing site issue described by Green et al pertains to KK lines only and does NOT affect GD or shRNA lines.
KK RNAi lines – landing site genotyping
All KK lines were genotyped using a simple diagnostic PCR, as described in Green et al., Nat. Methods (2014), to verify whether they contain a transgene insertion at the 30B and/or 40D landing site(s).
The genotyping results for all KK lines can be found here: All KK lines - 30B and 40D landing site insertion status
Disclaimer: Since the diagnostic PCR reactions were carried out just once per line, we do not guarantee that they are 100% accurate.
KK Protocols
- KK library cloning strategy
- KK library landing site
Annotated insertion: landing site VIE-260B (position chr2L: 22019296, cytological band 40D3) and a previously non-annotated insertion: (position chr2L: 9437482, cytological band 30B3). See KK library - landing site information for more details. - Bacteria Transformation
- Transgene Verification
- pKC26 transformation vector [MAP] [PDF] [TEXT]
- pKC43 landing site [MAP] [PDF] [TEXT]
shRNA Transgenic RNAi Library
The Vienna Drosophila Resource Center has created a small number of UAS-RNAi lines using short hairpin microRNA (shRNA) technology to extend and complement the current VDRC RNAi collection. 372 shRNA lines are available as of Jan 2017 and more are in our production pipeline.
Rationale
For commonly studied genes, where there is only 1 RNAi line in the VDRC GD or KK collection at present, we aim to add a further functional RNAi line to facilitate verification of phenotypes. We chose to use the short hairpin RNAi technology as it is a simpler and more cost-effective method of creating lines than by using long double-stranded RNA. Short hairpins RNAs (shRNAs), containing a 21bp targeting sequence embedded into a micro-RNA (miR-1) backbone, have been shown to be very effective for gene knockdown in both the germline and somatic tissues (Ni et al., 2011). This technology has been used extensively by the Transgenic RNAi Project (TRiP).
To avoid direct duplication of community resources, the VDRC has collaborated with the TRiP team during shRNA design to ensure that the new VDRC lines are as distinct as possible from the TRiP resource. We have used the WALIUM20 vector (for triggering RNAi in soma and germline) in combination with the attP40 landing site, meaning that both the chromosome and insertion site differ from the majority of TRiP lines, but the insertion chromosome (2nd) and selectable marker (white) are the same as the VDRC KK RNAi collection. In addition, the hairpin sequences we have selected target a different region of the mRNA sequence and are independent of all current resources. The new lines therefore extend the range of RNAi lines available to the Drosophila research community and are complementary to the RNAi resources currently available from the VDRC, Bloomington Drosophila Stock Center (BDSC), National Institute of Genetics (NIG) and TsingHua Fly Center (THFC).
Production pipeline overview
- Design possible 21mers, map SNPs and select best shRNA sequence.
- Anneal top and bottom oligos; ligate into pWALIUM20 vector.
- Confirm sequence of individual constructs.
- Combine 50-100 constructs and inject as a pool into w- embryos for targeted phiC31-mediated integration.
- Select w+ transformants and balance transgenic line.
- Sequence transgenic line to confirm identity of shRNA.
- Make line available via VDRC online ordering system.
| VDRC shRNA lines | ||
|---|---|---|
| Vector | pWALIUM20 (Perkins et al., 2015) map and sequence |
|
| Landing site | attP40 (Markstein et al., 2008) | |
| Integration locus | 2nd chromosome @ 25C6 | |
| Selectable marker | white | |
| Knock-down in | soma and germline | |
shRNA Library - detailed information
Hairpin design
- 21nt siRNA oligos were designed to target exons common to all isoforms of a gene using the DSIR website tool (http://biodev.extra.cea.fr/DSIR/DSIR.html) and an in-house off-target analysis script (Thomas Burkard, IMP/IMBA, Vienna).
- SNPs were called with GATK tool (Broad Inst.) using whole genome resequencing data from 10 Drosophila samples, aligned against the UCSC dm3 genome assembly.
- Overlap of a shRNA with a SNP was detected with an in-house script (BioComp, Vienna Biocenter Core Facilities).
- For each gene, the best shRNA sequence was selected based on a combination of highest predicted activity, lowest off-target probability and absence of common SNPs in a critical position.
- A small number (5%) of shRNAs exactly matching those already created or planned by TRiP (pers. comm. L.Perkins, C. Yu) were eliminated to avoid duplication of resources.
Construct generation
71 base oligos were designed according to the methods used by the TRiP team by concatenating the following:
Top strand oligo = 'ctagcagt', the sense-strand oligo (21 base pairs), 'tagttatattcaagcata', the anti-sense oligo, and 'gcg'.
Bottom strand oligo = 'aattcgc', the sense-strand oligo, 'tatgcttgaatataacta', the anti-sense oligo, and 'actg'.
Top and bottom 71mer oligos were annealed, creating overhangs for ligating into pWALIUM20 via the NheI and EcoRI sites, as described by Ni et al., 2011.
Ligation reactions were transformed into NEB10beta competent cells followed by colony PCR and sequencing to identify clones with the correct shRNA sequence.
PCR primers for confirming shRNA sequence:
fwd: 5'-ACCAGCAACCAAGTAAATCAAC-3'
rev: 5'-TAATCGTGTGTGATGCCTACC-3'
Sequencing primer:
5'-ACCAGCAACCAAGTAAATCAAC-3'
Only those constructs confirmed to have correct DNA were used for injection. Plasmid DNA was prepared from minipreps using GeneJET columns (Fisher Scientific). Typically, 50-100 constructs were pooled at equal concentrations, purified with AMPure XP beads (Agencourt) and resuspended at 200-250ng/ul.
Landing site
All fly shRNA stocks created by the VDRC are integrated into the attP40 genomic landing site on the left arm of the second chromosome at 25C6. The selectable marker is white, so the presence of the UAS-shRNA transgene can be confirmed by the stock having red eyes.
The attP40 site was chosen because it has been shown to provide high levels of induced expression of transgenes, yet maintain a low basal expression when the transgenes are not induced (Markstein et al., 2008).
The majority of the TRiP RNAi stocks have insertions in the attP2 genomic landing site on chromosome 3 with vermilion as the selectable marker, whereas the VDRC shRNA and KK lines have the RNAi construct inserted on chromosome 2 with white as the selectable marker.
Injections and crossing scheme
Since only one attB insert can integrate into an attP site via phiC31-mediated recombination (Groth et al., 2004), we injected constructs in pools of 50-100. Constructs were microinjected into embryos derived from a cross between males homozygous for the attP40 integration site (Markstein et al., 2008) and females homozygous for a fusion gene encoding the PhiC31 integrase under the control of the vasa promoter (vas-integ), which provides a maternal source of integrase (Bischof et al., 2007). Single males derived from these embryos were crossed to w- females, and males carrying the inserted construct (identified by their w+ eye colour) were selected; note that the source of integrase is also removed in this step. These w+ males were crossed to w;Sp/CyO females to establish balanced, homozygous stocks. The majority of stocks are homozygous viable, but a small number are homozygous sterile or lethal and still have CyO present (status listed under 'Viability' for each stock).
For crossing scheme figure see here.
Stock characterization and verification
In order to identify which of the pooled constructs were inserted in each transgenic line, genomic DNA was extracted as soon as w+ flies could be sacrificed, either directly after setting up the cross to the Sp/CyO balancer stock, or during the subsequent crosses to create a homozygous stock. The inserted DNA was amplified across the entire hairpin and sequenced to assign the stock to a construct.
PCR primers for confirming shRNA sequence:
fwd: 5'-ACCAGCAACCAAGTAAATCAAC-3'
rev: 5'-TAATCGTGTGTGATGCCTACC-3'
Sequencing primer:
5'-ACCAGCAACCAAGTAAATCAAC-3'
After genetic crosses to balance the stock and make it homozygous, the sequence of the transgene was again verified (using the same primers) before making the completed stock available here at www.vdrc.at.
Control lines
Currently, the best control for the shRNA lines is VDRC_ID 60200, containing an empty pWALIUM20 vector (no hairpin sequence) integrated into the attP40 landing site, created in the same way and with same gene genetic background as the shRNA lines. It can be found under 'Browse RNAi Toolkit Stocks'. We are creating an shRNA-EGFP line in the same way to provide a further control. Alternatively, we suggest using a VDRC shRNA line which is known NOT to function in the process or pathway being studied.
Acknowledgements
We thank the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing the pWALIUM20 plasmid vector used in the creation of the VDRC shRNA lines.
We are especially grateful to Liz Perkins, Claire Hu and Norbert Perrimon (TRiP, Harvard Medical School) for their helpful discussions and for coordinating the design of the shRNAs.
Please acknowledge the VDRC for providing the lines!
How transgenic RNAi works in Drosophila
The RNA interference (RNAi) pathway is often exploited in experimental biology to study the function of genes in cell culture and in vivo in model organisms. Double-stranded RNA is synthesized with a sequence complementary to a gene of interest and introduced into a cell or organism, where it is recognized as exogenous genetic material and activates the RNAi pathway. This leads to sequence-specific degradation of the target mRNA and thereby a drastic decrease in the expression of a targeted gene. Studying the effects of this decrease can show the physiological role of the gene product. Since RNAi may not totally abolish expression of the gene, this technique is referred to as a "knockdown", to distinguish it from "knockout" procedures in which expression of a gene is entirely eliminated.
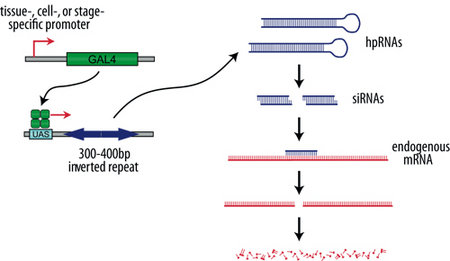
Figure 1: Transgenic RNAi in Drosophila. The generic GAL4/UAS system is used to drive the expression of a hairpin RNA (hpRNAs). These double-stranded RNAs are processed by Dicer into siRNAs which direct sequence-specific degradation of the target mRNA.
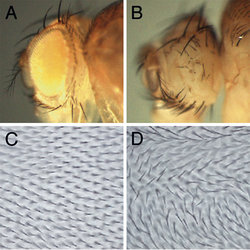
Figure 2: RNAi phenotypes. (A). Control with GAL4 driver only. (B). GAL4 driver + UAS-eyRNAi, targeting the eyeless gene. The eye is missing, as in the eyeless mutant. (C). Wing hairs in a wild-type fly all point in the same direction. (D). GAL4 driver + UAS-fmiRNAi, targeting flamingo, a gene required for planar cell polarity. The wing hairs are misorientated, as in the flamingo mutant.
The VDRC genome-wide Drosophila RNAi resource enables large scale genetic screens, making it possible to carry out loss-of-function experiments in essentially any tissue or cell at any stage in the life of the organism.
When crossed to a Gal4 'driver' line, the UAS-RNAi stocks induce expression of a specific hairpin structure, which silences expression of the target gene via RNA interference (RNAi).
The VDRC RNAi resource includes transgenic UAS-RNAi stocks with long hairpins (GD and KK libraries) and short hairpin microRNAs (shRNA library).
- Collectively, the RNAi lines cover a total of 12,671 (91%) of the D.melanogaster protein-coding genes.
- GD collection covers 11,292 genes (81%).
- KK collection covers 9,646 genes (69%).
- shRNA collection covers 372 genes (2.7%) - more under construction.
- For over 8500 genes, we have 2 or more independent RNAi lines available.
In addition, the VDRC provides:
- 13,848 DNA constructs used to generate the GD collection.
- Toolkit stocks used for construction of all 3 RNAi libraries.
RNAi Libraries Overview
Collectively, the RNAi lines cover a total of 12,671 (91%) of the D.melanogaster protein-coding genes. For over 8500 genes, we have 2 or more independent RNAi lines available.
| P-element library (GD) | ΦC31 library (KK) | Short hairpin microRNA library (shRNA) | |
|---|---|---|---|
| Available since | 2007 | 2009 | 2016 |
| Insertion of the UAS-IR | Random. Insertions mapped to chromosome X, 2 or 3. Precise locations unknown. | Targeted into VIE-260b landing site, chromosome 2, at position 22019296 (5' to CR33987) (see KK library - landing site information) | Targeted into attP40 landing site (Markstein et al., 2008), chromosome 2 at 25C6 |
| Target selection/ PCR template design | Dietzl et al (2007) Nature v448 p151 | Horn and Boutros (DKFZ, Heidelberg) | DSIR website tool and off-target analysis script (Burkard, IMP/IMBA, Vienna) |
| Hairpin length range/average (bp) | 109-400 (321) | 81-799 (357) | 21 |
| Transformation vector | pMF3 (10x UAS) | pKC26 (10x UAS) | pWALIUM20 (Perkins et al., 2015) |
| Verification of transformant lines | Restriction digest | Sanger sequencing | Sanger sequencing |
| Coverage | 16,763 lines 11,292 genes (81%) Average of 1.19 lines/gene |
9,822 lines 9,646 genes (69%) Average of 0.7 lines/gene |
372 lines (as of Jan 2017). More to be added. |
| Activity | Soma and germline. Enhanced by co-expression of Dicer2 | Soma and germline. Enhanced by co-expression of Dicer2 | Soma and germline. |
GD Transgenic RNAi Library
The GD library comprises 16,763 transgenic Drosophila strains, each containing an inducible UAS-RNAi construct against a single protein-coding gene. All insertions have been molecularly validated, and a sample also functionally validated. We estimate that >80% of the lines provide potent and gene-specific silencing.
GD library cloning strategy
The library was constructed by cloning gene fragments of 300-400bp as inverted repeats (IRs) in the antisense-sense orientation into a modified pUAST vector pMF3 with 10 copies of UAS sites (see map). To amplify 300-400bp gene fragments, primers were designed to amplify from every predicted protein-coding gene in Release 4.3 of the Drosophila genome sequence. 15,072 PCR products were successfully amplified and cloned as IRs into pMF3, representing 13,344 genes or 97% of the Drosophila genome. The GD library insertions are P-element based transgenes with random insertion sites. Insertions have been mapped to a chromosome (1, 2, or 3) but we do not have data on specific location on the chromosome.
Publication showing creation and validation of GD library:
- A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila.
Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ.
Nature. 2007 Jul 12;448(7150):151-6.
PMID: 17625558
[Abstract]
GD Protocols
- GD library cloning strategy
- Genomic DNA extraction
- mRNA extraction
- 1st strand cDNA
- 96 well cloning of RNAi constructs
- 96 well DNA purification using 'SigmaSpin Post-Reaction Clean-Up Plates'
- 96 well DNA gel extraction using 'NucleoSpin Multi-96 Extract'
- Fly transformation
- Bacteria Transformation
- Transgene Verification
- pMF3 transformation vector [MAP] [PDF] [TEXT]
KK Transgenic RNAi Library
The KK library, created after the GD library, is a genome-wide transgenic RNAi library.
The main motivation for generating a new library was to:
- improve consistency and viability by integrating all constructs at the same landing site
- increase efficiency and specificity of gene disruption by using improved Inverted Repeat (IR) design
- generate a line for genes not covered by the GD library
- provide an independent construct to verify phenotypic effects.
A key difference between the GD and KK libraries is that the GD library insertions are P-element based transgenes with random insertion sites, whereas the KK library contains phiC31-based transgenes with a single, defined insertion site.
The KK library currently comprises 9,822 transgenic Drosophila strains, each containing an inducible UAS-RNAi construct against a single protein-coding gene. The sequence of all insertions have been validated by sequencing, and a sample also functionally validated. Based on an initial benchmark study of 164 genes with known lethality/morphological phenotypes, >90% of the KK lines provided potent and gene-specific silencing when crossed to the ubiquitous driver Act5C-GAL4 (Keleman et al., unpublished).
KK library - landing site information
KK RNAi lines landing site - background information
During construction of the KK RNAi library, the hairpin constructs were targeted to a single genomic location. However, Green et al. (2014) have previously shown that there are actually 2 potential genomic landing sites for the KK RNAi constructs:
- At 40D3 (the originally intended VIE-260B landing site, in the 5'UTR of the gene tiptop, position chr2L: 22019296)
- At 30B3 (the actual site, position chr2L: 9437482).
In 75% of the KK lines, the insertion of the RNAi hairpin construct is in the 30B landing site only and there are no problems. However, in ~25% of KK lines there is an additional insertion of the RNAi construct at the 40D landing site and this causes non-specific phenotypes when crossed to some Gal4 drivers. See Green et al., Nat. Methods (2014) for details.
A publication investigating this issue further, from the lab of Kieran Harvey (Vissers et al., Nat. Comm. (2016)), confirms that approximately 25% of VDRC KK RNAi lines (38 out of 150) can cause false-positive phenotypes. Whilst working on the hippo pathway and performing screens in the eye, Vissers et al. noticed phenotypes which were due to the insertion site and not due to expression of the RNAi hairpin. They were able to attribute the false-positive phenotypes to enhancement of the Hippo pathway, owing to ectopic UAS-driven expression of the Tiptop (tio) transcription factor - a consequence of the 40D landing site being located in the tio 5' UTR.
KK library control lines
As a control line for experiments involving KK lines, the VDRC provides the host strain for the KK library (VDRC_ID 60100). This line contains landing sites at both 40D and 30B.
A number of additional control lines for the KK library, donated by Kieran Harvey (Peter MacCallum Cancer Centre, Melbourne, Australia) are available. (For details see Vissers et al., (2016): A Drosophila RNAi library modulates Hippo pathway-dependent tissue growth).
The collection consists of 5 lines which can be displayed by using the search term "....harvey".
An RNAi-toolkit stock, 40D-UAS (VDRC_ID 60101) where the landing sites at 40D and 30B in the KK library host strain have been separated by meiotic recombination. This line contains Gal4-responsive UAS repeats but no functional RNAi coding sequence at 40D and no transgene insertion at 30B and enables researchers to easily test, in a single cross, whether their genetic screen of interest will be affected by ectopic tiptop expression.
4 further KK lines for specific genes were created by meiotic recombination to separate the insertions at 30B and 40D. These lines contain an insertion of the UAS-RNAi construct in the landing site at 30B only and no insertion at the 40D landing site.
- VDRC_ID 111000 is the '30D only' version of VDRC_ID 107151 (CG8208, MBD-like)
- VDRC_ID 111001 is the '30D only' version of VDRC_ID 104523 (CG4005, Yki)
- VDRC_ID 111002 is the '30D only' version of VDRC_ID 106174 (CG12072, Wts)
- VDRC_ID 111003 is the '30D only' version of VDRC_ID 105838 (CG4236, Caf1)
When using these additional control lines, please reference Vissers et al (2016) and acknowledge the VDRC for providing the lines.
We would like to emphasise that ~75% of KK lines (insertion at 30D only) are absolutely fine and that the remaining 25% (insertion at 40D) will only be a problem in cases where an assay is affected by misexpression of the tio gene. If you are planning to use KK lines and are concerned that you will see non-specific phenotypes, we suggest that you cross your Gal4 driver of interest to the 40D-UAS line and check whether the F1 progeny demonstrate a dominant phenotype. If your assay is not affected by tio misexpression, you can use all 9,822 KK lines with confidence, but if you see an effect, then 25% of the KK lines will give misleading results in that particular screen.
Validation of KK lines
The VDRC have shipped more than 400,000 KK lines to the Drosophila research community. We would like to reassure researchers that use of KK lines has resulted in several hundred publications in which the KK line phenotypes have been validated with independent RNAi lines targeting a different region of the same gene and with other insertion sites. These publications include some large scale screens, e.g.:
- Czech B et al., (2013). A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol Cell. 2013 Jun 6;50(5):749-61.
[PubMed]. - Berns N et al., (2014). A genome-scale in vivo RNAi analysis of epithelial development in Drosophila identifies new proliferation domains outside of the stem cell niche. J Cell Sci. 2014 Jun 15;127(Pt 12):2736-48.
[PubMed]. - Homem CC et al., (2014). Ecdysone and mediator change energy metabolism to terminate proliferation in Drosophila neural stem cells. Cell. 2014 Aug 14;158(4):874-88.
[PubMed].
Verify your results!
As is recommended for results obtained using any RNAi lines, we advise verifying your observed KK phenotype with an alternative RNAi line containing an independent RNAi hairpin construct targeting the same gene and a different genomic insertion site where possible.
Please note that the GD library was generated using P-element insertion and the shRNA library transgenes are inserted in the attP40 landing site, therefore the landing site issue described by Green et al pertains to KK lines only and does NOT affect GD or shRNA lines.
KK RNAi lines – landing site genotyping
All KK lines were genotyped using a simple diagnostic PCR, as described in Green et al., Nat. Methods (2014), to verify whether they contain a transgene insertion at the 30B and/or 40D landing site(s).
The genotyping results for all KK lines can be found here: All KK lines - 30B and 40D landing site insertion status
Disclaimer: Since the diagnostic PCR reactions were carried out just once per line, we do not guarantee that they are 100% accurate.
KK Protocols
- KK library cloning strategy
- KK library landing site
Annotated insertion: landing site VIE-260B (position chr2L: 22019296, cytological band 40D3) and a previously non-annotated insertion: (position chr2L: 9437482, cytological band 30B3). See KK library - landing site information for more details. - Bacteria Transformation
- Transgene Verification
- pKC26 transformation vector [MAP] [PDF] [TEXT]
- pKC43 landing site [MAP] [PDF] [TEXT]
shRNA Transgenic RNAi Library
The Vienna Drosophila Resource Center has created a small number of UAS-RNAi lines using short hairpin microRNA (shRNA) technology to extend and complement the current VDRC RNAi collection. 372 shRNA lines are available as of Jan 2017 and more are in our production pipeline.
Rationale
For commonly studied genes, where there is only 1 RNAi line in the VDRC GD or KK collection at present, we aim to add a further functional RNAi line to facilitate verification of phenotypes. We chose to use the short hairpin RNAi technology as it is a simpler and more cost-effective method of creating lines than by using long double-stranded RNA. Short hairpins RNAs (shRNAs), containing a 21bp targeting sequence embedded into a micro-RNA (miR-1) backbone, have been shown to be very effective for gene knockdown in both the germline and somatic tissues (Ni et al., 2011). This technology has been used extensively by the Transgenic RNAi Project (TRiP).
To avoid direct duplication of community resources, the VDRC has collaborated with the TRiP team during shRNA design to ensure that the new VDRC lines are as distinct as possible from the TRiP resource. We have used the WALIUM20 vector (for triggering RNAi in soma and germline) in combination with the attP40 landing site, meaning that both the chromosome and insertion site differ from the majority of TRiP lines, but the insertion chromosome (2nd) and selectable marker (white) are the same as the VDRC KK RNAi collection. In addition, the hairpin sequences we have selected target a different region of the mRNA sequence and are independent of all current resources. The new lines therefore extend the range of RNAi lines available to the Drosophila research community and are complementary to the RNAi resources currently available from the VDRC, Bloomington Drosophila Stock Center (BDSC), National Institute of Genetics (NIG) and TsingHua Fly Center (THFC).
Production pipeline overview
- Design possible 21mers, map SNPs and select best shRNA sequence.
- Anneal top and bottom oligos; ligate into pWALIUM20 vector.
- Confirm sequence of individual constructs.
- Combine 50-100 constructs and inject as a pool into w- embryos for targeted phiC31-mediated integration.
- Select w+ transformants and balance transgenic line.
- Sequence transgenic line to confirm identity of shRNA.
- Make line available via VDRC online ordering system.
| VDRC shRNA lines | ||
|---|---|---|
| Vector | pWALIUM20 (Perkins et al., 2015) map and sequence |
|
| Landing site | attP40 (Markstein et al., 2008) | |
| Integration locus | 2nd chromosome @ 25C6 | |
| Selectable marker | white | |
| Knock-down in | soma and germline | |
shRNA Library - detailed information
Hairpin design
- 21nt siRNA oligos were designed to target exons common to all isoforms of a gene using the DSIR website tool (http://biodev.extra.cea.fr/DSIR/DSIR.html) and an in-house off-target analysis script (Thomas Burkard, IMP/IMBA, Vienna).
- SNPs were called with GATK tool (Broad Inst.) using whole genome resequencing data from 10 Drosophila samples, aligned against the UCSC dm3 genome assembly.
- Overlap of a shRNA with a SNP was detected with an in-house script (BioComp, Vienna Biocenter Core Facilities).
- For each gene, the best shRNA sequence was selected based on a combination of highest predicted activity, lowest off-target probability and absence of common SNPs in a critical position.
- A small number (5%) of shRNAs exactly matching those already created or planned by TRiP (pers. comm. L.Perkins, C. Yu) were eliminated to avoid duplication of resources.
Construct generation
71 base oligos were designed according to the methods used by the TRiP team by concatenating the following:
Top strand oligo = 'ctagcagt', the sense-strand oligo (21 base pairs), 'tagttatattcaagcata', the anti-sense oligo, and 'gcg'.
Bottom strand oligo = 'aattcgc', the sense-strand oligo, 'tatgcttgaatataacta', the anti-sense oligo, and 'actg'.
Top and bottom 71mer oligos were annealed, creating overhangs for ligating into pWALIUM20 via the NheI and EcoRI sites, as described by Ni et al., 2011.
Ligation reactions were transformed into NEB10beta competent cells followed by colony PCR and sequencing to identify clones with the correct shRNA sequence.
PCR primers for confirming shRNA sequence:
fwd: 5'-ACCAGCAACCAAGTAAATCAAC-3'
rev: 5'-TAATCGTGTGTGATGCCTACC-3'
Sequencing primer:
5'-ACCAGCAACCAAGTAAATCAAC-3'
Only those constructs confirmed to have correct DNA were used for injection. Plasmid DNA was prepared from minipreps using GeneJET columns (Fisher Scientific). Typically, 50-100 constructs were pooled at equal concentrations, purified with AMPure XP beads (Agencourt) and resuspended at 200-250ng/ul.
Landing site
All fly shRNA stocks created by the VDRC are integrated into the attP40 genomic landing site on the left arm of the second chromosome at 25C6. The selectable marker is white, so the presence of the UAS-shRNA transgene can be confirmed by the stock having red eyes.
The attP40 site was chosen because it has been shown to provide high levels of induced expression of transgenes, yet maintain a low basal expression when the transgenes are not induced (Markstein et al., 2008).
The majority of the TRiP RNAi stocks have insertions in the attP2 genomic landing site on chromosome 3 with vermilion as the selectable marker, whereas the VDRC shRNA and KK lines have the RNAi construct inserted on chromosome 2 with white as the selectable marker.
Injections and crossing scheme
Since only one attB insert can integrate into an attP site via phiC31-mediated recombination (Groth et al., 2004), we injected constructs in pools of 50-100. Constructs were microinjected into embryos derived from a cross between males homozygous for the attP40 integration site (Markstein et al., 2008) and females homozygous for a fusion gene encoding the PhiC31 integrase under the control of the vasa promoter (vas-integ), which provides a maternal source of integrase (Bischof et al., 2007). Single males derived from these embryos were crossed to w- females, and males carrying the inserted construct (identified by their w+ eye colour) were selected; note that the source of integrase is also removed in this step. These w+ males were crossed to w;Sp/CyO females to establish balanced, homozygous stocks. The majority of stocks are homozygous viable, but a small number are homozygous sterile or lethal and still have CyO present (status listed under 'Viability' for each stock).
For crossing scheme figure see here.
Stock characterization and verification
In order to identify which of the pooled constructs were inserted in each transgenic line, genomic DNA was extracted as soon as w+ flies could be sacrificed, either directly after setting up the cross to the Sp/CyO balancer stock, or during the subsequent crosses to create a homozygous stock. The inserted DNA was amplified across the entire hairpin and sequenced to assign the stock to a construct.
PCR primers for confirming shRNA sequence:
fwd: 5'-ACCAGCAACCAAGTAAATCAAC-3'
rev: 5'-TAATCGTGTGTGATGCCTACC-3'
Sequencing primer:
5'-ACCAGCAACCAAGTAAATCAAC-3'
After genetic crosses to balance the stock and make it homozygous, the sequence of the transgene was again verified (using the same primers) before making the completed stock available here at www.vdrc.at.
Control lines
Currently, the best control for the shRNA lines is VDRC_ID 60200, containing an empty pWALIUM20 vector (no hairpin sequence) integrated into the attP40 landing site, created in the same way and with same gene genetic background as the shRNA lines. It can be found under 'Browse RNAi Toolkit Stocks'. We are creating an shRNA-EGFP line in the same way to provide a further control. Alternatively, we suggest using a VDRC shRNA line which is known NOT to function in the process or pathway being studied.
Acknowledgements
We thank the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing the pWALIUM20 plasmid vector used in the creation of the VDRC shRNA lines.
We are especially grateful to Liz Perkins, Claire Hu and Norbert Perrimon (TRiP, Harvard Medical School) for their helpful discussions and for coordinating the design of the shRNAs.
Please acknowledge the VDRC for providing the lines!
How transgenic RNAi works in Drosophila
The RNA interference (RNAi) pathway is often exploited in experimental biology to study the function of genes in cell culture and in vivo in model organisms. Double-stranded RNA is synthesized with a sequence complementary to a gene of interest and introduced into a cell or organism, where it is recognized as exogenous genetic material and activates the RNAi pathway. This leads to sequence-specific degradation of the target mRNA and thereby a drastic decrease in the expression of a targeted gene. Studying the effects of this decrease can show the physiological role of the gene product. Since RNAi may not totally abolish expression of the gene, this technique is referred to as a "knockdown", to distinguish it from "knockout" procedures in which expression of a gene is entirely eliminated.

Figure 1: Transgenic RNAi in Drosophila. The generic GAL4/UAS system is used to drive the expression of a hairpin RNA (hpRNAs). These double-stranded RNAs are processed by Dicer into siRNAs which direct sequence-specific degradation of the target mRNA.

Figure 2: RNAi phenotypes. (A). Control with GAL4 driver only. (B). GAL4 driver + UAS-eyRNAi, targeting the eyeless gene. The eye is missing, as in the eyeless mutant. (C). Wing hairs in a wild-type fly all point in the same direction. (D). GAL4 driver + UAS-fmiRNAi, targeting flamingo, a gene required for planar cell polarity. The wing hairs are misorientated, as in the flamingo mutant.
